RESEARCH
Understanding how cells grow and divide has profound impacts on basic science, biotechnology, and medicine. Despite recent advances in molecular biology and biochemistry, a central challenge remains: bridging the nanometer-scale activities of proteins and the construction of entire cells. Although the mechanisms of bacterial proliferation have been a major focus of research for over a century, it has remained difficult to determine how cellular structure and organization are dynamically controlled due to the central—yet neglected—importance of physical factors.
To address these knowledge gaps, our lab pursues research directions that span from the atomic to the multicellular scales. We investigate the physical nature of intracellular spatial organization, mechanics, and kinetics by leveraging top-down approaches based on cellular-scale observations, bottom-up approaches based on biophysical molecular observations, and computational modeling that connects the two paradigms. Understanding cellular growth and form remains a fascinating, multifaceted challenge with obvious implications for health and disease. In addition to the importance of bacteria as a model system for basic science, uncovering the general physical rules that underlie how bacteria grow and divide will have important applications for controlling bacterial communities and developing novel strategies in synthetic biology.
| Research Topics |
|---|
| Cultivation of in vitro communities to model the microbiota |
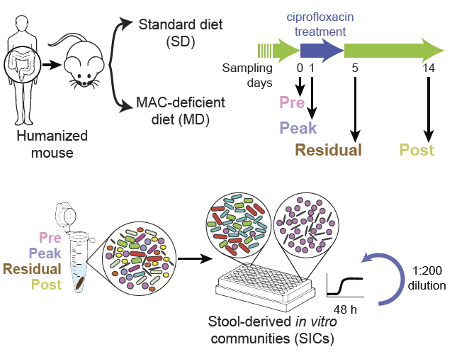
The gut microbiota, the dense and diverse community of microbes that occupy the gastrointestinal tract, perform many important functions for host physiology, including metabolism, xenobiotic processing, and protection against pathogen invasion. Mechanistic understanding of the gut-associate microbiotas has been hampered by limited throughput. To enable systematic, high-throughput interrogation of human-relevant microbial communities, we generate hundreds of in vitro communities cultured from diverse samples throughout the gastrointestinal tract in different media. These in vitro communities are phylogenetically complex, diverse, stable, and highly reproducible. They serve as a powerful resource for understanding species-species interactions in microbiota research.
High-throughput cultivation of stable, diverse, fecal-derived microbial communities to model the intestinal microbiota
Aranda-Diaz A, Ng K, Thomsen T, Real-Ramirez I, Dahan D, Dittmar S, Gonzalez CG, Chavez T, Vasquez K, Ngyuen T, Yu FB, Higginbottom SK, Neff NF, Elias JE, Sonnenburg JL, and Huang KC.
BioRxiv, (2020)
Bacterial interspecies interactions modulate pH-mediated antibiotic tolerance
Aranda-Diaz A, Obadia B, Dodge R, Thomsen T , Hallberg Z, Guvener Z, Ludington W, and Huang KC.
eLife, (2020)