RESEARCH
Understanding how cells grow and divide has profound impacts on basic science, biotechnology, and medicine. Despite recent advances in molecular biology and biochemistry, a central challenge remains: bridging the nanometer-scale activities of proteins and the construction of entire cells. Although the mechanisms of bacterial proliferation have been a major focus of research for over a century, it has remained difficult to determine how cellular structure and organization are dynamically controlled due to the central—yet neglected—importance of physical factors.
To address these knowledge gaps, our lab pursues research directions that span from the atomic to the multicellular scales. We investigate the physical nature of intracellular spatial organization, mechanics, and kinetics by leveraging top-down approaches based on cellular-scale observations, bottom-up approaches based on biophysical molecular observations, and computational modeling that connects the two paradigms. Understanding cellular growth and form remains a fascinating, multifaceted challenge with obvious implications for health and disease. In addition to the importance of bacteria as a model system for basic science, uncovering the general physical rules that underlie how bacteria grow and divide will have important applications for controlling bacterial communities and developing novel strategies in synthetic biology.
| Research Topics |
|---|
| The spatial organization of the gut microbiome |
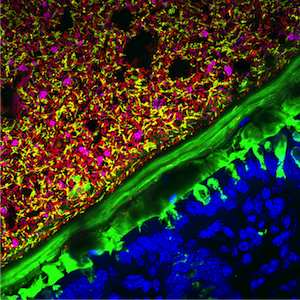 The community of microorganisms within our gastrointestinal tract, collectively termed the gut microbiota, constitutes one of the densest and most diverse bacterial ecosystems known. The complexity of this community presents unique challenges for studying the relationship between humans and their commensal microbes. Many gut microbiota studies focus on the enumeration of bacterial composition. However, measurements of bulk populations provide a low-resolution view of the interactions among microbes, host cells, and the local environment within the dynamic and heterogeneous intestinal ecosystem. Heterogeneous spatial organization is the norm in natural microbial ecosystems ranging from deep-sea methane vents to the surface of thermophilic sludge granules. The organizational principles that govern our internal ecosystem remain poorly understood, despite emerging ecological paradigms that have implications for localization of microbes within the gut. To address this shortcoming, in a close collaboration with the Sonnenburg lab we have developed a pipeline for the assessment of intestinal microbiota localization within immunofluorescence images of fixed gut cross-sections. The pipeline includes a flexible software package, BacSpace, for high-throughput quantification of microbial organization, including proximity to host mucus and epithelium. Using gnotobiotic and human microbiota-colonized (humanized) mice, we have demonstrated that elimination of microbiota accessible carbohydrates (MACs) from the diet, which is known to increase microbiota utilization of mucosal glycans, results in thinner mucus in the distal colon, increased proximity of microbes to the epithelium, and heightened expression of the inflammatory marker REG3β. This broadly applicable framework should accelerate the elucidation of the roles of microbiota localization in health and disease.
The community of microorganisms within our gastrointestinal tract, collectively termed the gut microbiota, constitutes one of the densest and most diverse bacterial ecosystems known. The complexity of this community presents unique challenges for studying the relationship between humans and their commensal microbes. Many gut microbiota studies focus on the enumeration of bacterial composition. However, measurements of bulk populations provide a low-resolution view of the interactions among microbes, host cells, and the local environment within the dynamic and heterogeneous intestinal ecosystem. Heterogeneous spatial organization is the norm in natural microbial ecosystems ranging from deep-sea methane vents to the surface of thermophilic sludge granules. The organizational principles that govern our internal ecosystem remain poorly understood, despite emerging ecological paradigms that have implications for localization of microbes within the gut. To address this shortcoming, in a close collaboration with the Sonnenburg lab we have developed a pipeline for the assessment of intestinal microbiota localization within immunofluorescence images of fixed gut cross-sections. The pipeline includes a flexible software package, BacSpace, for high-throughput quantification of microbial organization, including proximity to host mucus and epithelium. Using gnotobiotic and human microbiota-colonized (humanized) mice, we have demonstrated that elimination of microbiota accessible carbohydrates (MACs) from the diet, which is known to increase microbiota utilization of mucosal glycans, results in thinner mucus in the distal colon, increased proximity of microbes to the epithelium, and heightened expression of the inflammatory marker REG3β. This broadly applicable framework should accelerate the elucidation of the roles of microbiota localization in health and disease.
The effect of microbial colonization on the host proteome varies by gastrointestinal location.
Lichtman JS, Alsentzer E, Jaffe M, Sprockett D, Masutani E, Ikwa E, Fragiadakis GK, Clifford D, Huang BE, Sonnenburg JL†, Huang KC†, Elias JE†.
ISME J. 2016 May;10(5):1170-81. doi: 10.1038/ismej.2015.187. Epub 2015 Nov 17.
†Co-corresponding authors
Quantitative Imaging of Gut Microbiota Spatial Organization.
Earle KA*, Billings G*, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, Sonnenburg JL.
Cell Host Microbe. 2015 Oct 14;18(4):478-88. doi: 10.1016/j.chom.2015.09.002. Epub 2015 Oct 1.
*Co-first authors