RESEARCH
Understanding how cells grow and divide has profound impacts on basic science, biotechnology, and medicine. Despite recent advances in molecular biology and biochemistry, a central challenge remains: bridging the nanometer-scale activities of proteins and the construction of entire cells. Although the mechanisms of bacterial proliferation have been a major focus of research for over a century, it has remained difficult to determine how cellular structure and organization are dynamically controlled due to the central—yet neglected—importance of physical factors.
To address these knowledge gaps, our lab pursues research directions that span from the atomic to the multicellular scales. We investigate the physical nature of intracellular spatial organization, mechanics, and kinetics by leveraging top-down approaches based on cellular-scale observations, bottom-up approaches based on biophysical molecular observations, and computational modeling that connects the two paradigms. Understanding cellular growth and form remains a fascinating, multifaceted challenge with obvious implications for health and disease. In addition to the importance of bacteria as a model system for basic science, uncovering the general physical rules that underlie how bacteria grow and divide will have important applications for controlling bacterial communities and developing novel strategies in synthetic biology.
| Research Topics |
|---|
| Single-molecule imaging in live bacteria |
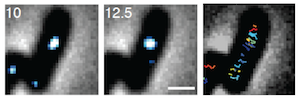 Although protein complexes coordinate the activities of many enzymes in space and time, it was previously unknown whether growth of the bacterial cell wall involves stable synthetic complexes. We used single-molecule tracking to demonstrate that the highly dynamic association between an essential cell-wall synthesis enzyme and the cytoskeleton allows the cell to buffer growth rate against large fluctuations in enzyme abundance (Lee et al., 2014). Importantly, dynamic association is thus an efficient strategy for coordinating multiple enzymes, suggesting novel and counterintuitive strategies for interfering with function by stabilizing interactions, for example during pathogenesis.
Although protein complexes coordinate the activities of many enzymes in space and time, it was previously unknown whether growth of the bacterial cell wall involves stable synthetic complexes. We used single-molecule tracking to demonstrate that the highly dynamic association between an essential cell-wall synthesis enzyme and the cytoskeleton allows the cell to buffer growth rate against large fluctuations in enzyme abundance (Lee et al., 2014). Importantly, dynamic association is thus an efficient strategy for coordinating multiple enzymes, suggesting novel and counterintuitive strategies for interfering with function by stabilizing interactions, for example during pathogenesis.
Single molecule imaging reveals the in vivo regulation of the activity of bifunctional cell wall synthesis enzymes.
Lee TK, Meng K, Shi H, Huang KC, in press, Nat Communications.
A dynamically assembled cell wall synthesis machinery buffers cell growth.
Lee TK, Tropini C, Hsin J, Desmarais SM, Ursell TS, Gong E, Gitai Z, Monds RD†, Huang KC†.
Proc Natl Acad Sci U S A. 2014 Mar 25;111(12):4554-9. doi: 10.1073/pnas.1313826111. Epub 2014 Feb 18.
†Co-corresponding authors